Pandas for Large Data: Examples and Tips
Last updated:- Use small data types
- Delete objects from memory
- Do joins chunk by chunk
- Use vectorized numpy functions when possible
- Parallel apply with swifter
- Parallel dataframe processing with multiprocessing
WIP Alert This is a work in progress. Current information is correct but more content may be added in the future.
Use small data types
Use
df.dtypesto see what your dataframe dtypes look like
Pandas sometimes defaults to unnecessarily large datatypes.
Use the smallest dtypes you possibly can such as:
'uint8'instead of'int32'or'int64''float32'instead of'float64''category'instead'object'for categorical data
Delete objects from memory
Delete DataFrames and other variables you will not use anymore using:
del(my_df)deletes the reference to your variablegc.collect()explictly frees memory space.
Example:
import gc
import pandas as pd
df1 = load_data()
df1_processed = process_df(df1)
# df1 is not needed anymore
del(df1)
gc.collect()
# you'll have more memory to load other dataframes
df2 = load_data_2()
// ...
Do joins chunk by chunk
Perform large, complex joins in parts to avoid exploding memory usage:
# split dataframe indices into parts
indexes = np.linspace(0, len(second_df), num=10, dtype=np.int32)
# update in small portions
for i in range(len(indexes)-1):
my_df = pd.concat(
[
my_df, # the same DF
pd.merge(left=pd.merge(
left=second_df.loc[indexes[i]:indexes[i+1], :],
right=third_df,
how='left',
on='foreign_key'
),
right=fourth_df,
how='left',
on='other_foreign_key'
)
]
)
Use vectorized numpy functions when possible
For example, use DataFrame.apply instead of Series.map to create derived columns and/or to operate on columns:
Time comparison: create a dataframe with 10,000,000 rows and multiply a numeric column by 2
import pandas as pd
import numpy as np
# create a sample dataframe with 10,000,000 rows
df = pd.DataFrame({
'x': np.random.normal(loc=0.0, scale=1.0, size=10000000)
})
 Sample dataframe for benchmarking
Sample dataframe for benchmarking (top 5 rows shown only)
Using map function multiply 'x' column by 2
def multiply_by_two_map(x):
return x*2
df['2x_map'] = df['x'].map(multiply_by_two)
# >>> CPU times: user 14.4 s, sys: 300 ms, total: 14.7 s
# >>> Wall time: 14.7 s
Using apply function multiply 'x' column by 2
import numpy as np
def multiply_by_two(arr):
return np.multiply(arr,2)
# note the double square brackets around the 'x'!!
# this is because we want to use DataFrame.apply,
# not Series.apply!!
df['2x_apply'] = df[['x']].apply(multiply_by_two)
# >>> CPU times: user 80 ms, sys: 112 ms, total: 192 ms
# >>> Wall time: 188 ms
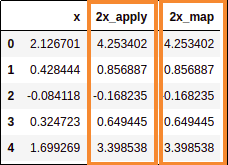 Both map and apply yielded the same results
Both map and apply yielded the same results but the apply version was around
70 times faster!
Parallel apply with swifter
see pandas column operations: map vs apply for a comparison between map and apply
The full comparison code is on this notebook
Swifter is a library that aims to parallelize Pandas apply whenever possible.
It is not always the case that using swifter is faster than a simple Series.
| Dataframe.apply | Series.apply | Dataframe.apply (Swifter) | Series.apply (swifter) | |
|---|---|---|---|---|
| Vectorizable operations | 0.5 secs | 6 secs | 0.3 secs | 0.2 secs |
| Number to string | 24 secs | 137 secs | ||
| Simple if-then-else | 4.5 secs | 3 secs |
Parallel dataframe processing with multiprocessing
All chunks must fit in memory at the same time!
Spawn multiple Python processes and have each of them process a chunk of a large dataframe.
This will reduce the processing time by half or even more, depending on the number of processe you use.
All code available on this jupyter notebook
Example: use 8 cores to process a text dataframe in parallel.
Step 1: put all processing logic into a single function:
import pandas as pd # this is an example of common operations you have to perform on text # data to go from raw text to clean text you can use for modelling def process_df_function(df): output_df = df.copy() # replace weird double quotes with normal ones output_df['text'] = output_df['text'].apply(lambda text: text.replace("``",'"')) # text to lower case output_df['text'] = output_df['text'].apply(lambda text: text.lower()) # replace number with a special token output_df['text'] = output_df['text'].apply(lambda text: re.sub(r"\b\d+\b","tok_num", text)) # get the number of words in each text output_df['num_words'] = output_df['text'].apply(lambda text: len(re.split(r"(?:\s+)|(?:,)|(?:\-)",text))) # take out texts that are too large indices_to_remove_too_large = output_df[output_df['num_words'] > 50] output_df.drop(indices_to_remove_too_large.index, inplace=True) # or too small indices_to_remove_too_small = output_df[output_df['num_words'] < 10] output_df.drop(indices_to_remove_too_small.index, inplace=True) output_df.reset_index(drop=True, inplace=True) return output_dfStep 2: Use
multiprocessing.Poolto distribute the work over multiple processesimport multiprocessing import numpy as np import pandas as pd # load raw data into a dataframe raw_df = load_data("path/to/dataset") NUM_CORES = 8 # split the raw dataframe into chunks df_chunks = np.array_split(raw_df ,NUM_CORES) # use a pool to spawn multiple proecsses with multiprocessing.Pool(NUM_CORES) as pool: # concatenate all processed chunks together. # process_df_function is the function you defined in the previous block processed_df = pd.concat(pool.map(process_df_function, df_chunks), ignore_index=True)